Introduction
Developing a biomedical ontology covering a specific domain (e.g., OBI for biomedical investigations, VO for vaccine) is often an ambitious project. OBO Foundry principles have been developed to ensure collaborative ontology development and interoperability among ontologies. Examples of these principles include: a) ontologies are developed in a collaborative effort, b) ontologies use common relations that are unambiguously defined, c) ontologies provide procedures for user feedback and for identifying successive versions, and d) ontologies have a clearly bounded subject-matter.
With the increased size of a reference ontology, it is desirable to specify the terms in the ontology or generate a subset of the ontology that are needed for a particular application based on the user's request. In addition, user-friendly labels for a specific community is also desired. Ontodog is developed for meeting these needs. The ontology community view can be generated using different approaches. Ontodog uses different annotation layers to specify the terms that are of interest by a particular user group and who provide user preferred labels. Various annotation layers can be imported to the source ontology or the subset of source ontology providing lots of flexibility. This approach guarantees changes are not made in the source ontology and can easily be reused by different users for different purposes. The structure of ontology community view is illustrated in the figure shown below.
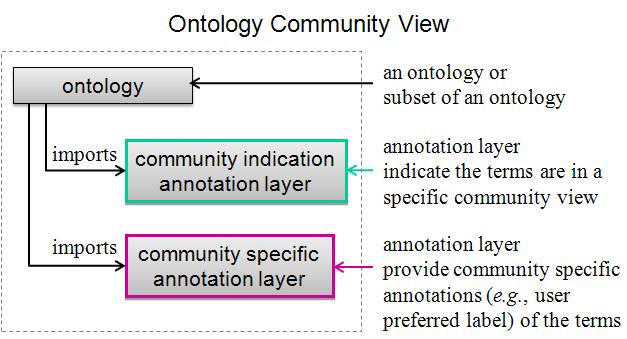
Ontodog is a web-based ontology community view generator. It can generate inSubset annotation ontology, user preferred label annotation ontology and subset of source ontology.
Ontodog performs the basic ontology modularization-like function, i.e.,it automatically extracts all axioms and related terms associated with user-specified signature term(s). In addition, Ontodog includes extra features: (1) extracting all instance data associated with the retrieved class terms and annotations; and (2) recursively extracting all axioms and related terms indirectly associated with signature terms. More features are being added to Ontodog, such as relabeling preferred names for various ontology terms to fit in with the needs from a specific community.
The Ontodog input data requires a source ontology and a list of user-specified signature terms in tab-delimited format. Ontodog provides the template files for generating the signature terms as the input terms file to download. There are several output options that the users can choose based on their needs.
With more and more ontologies being developed, Ontodog offers a timely web-based package of solutions for ontology view generation. Ontodog provides an efficient approach to promote ontology sharing and interoperability. It is easy to use and does not require knowledge of SPARQL, script programming, and command line operation. Ontodog is developed to serve the ontology community for ontology reuse. It is freely available under the Apache License 2.0.
Suggestions and comments are welcome. Thank you!
